Filtering GRIB data
[1]:
import time
import metview as mv
read() versus select()
Metview provides two ways to filter GRIB messages. Here is a quick comparison:
-
works with GRIB data from ECMWF’s archive, MARS
uses the same keywords as retrieve()
includes area cropping, spectral transforms and grid interpolation
has an equivalent icon in the graphical user interface, “GRIB Filter”
filtering may not be fully functional with non-MARS GRIB data
-
works with any GRIB data that ecCodes understands
uses ecCodes keys rather than MARS keywords
much faster than read() when called multiple times on the same Fieldset
no post-processing of data (use read() or regrid() separately for that)
only available from Python
[2]:
filename = "data_fc.grib"
if mv.exist(filename):
fs = mv.read(filename)
else:
fs = mv.gallery.load_dataset(filename)
Inspecting the data
A separate notebook covers the details of inspecting GRIB data with Metview. Here we will use the methods to decide which fields to filter.
[3]:
print(fs)
Fieldset (288 fields)
[4]:
fs.describe()
[4]:
| parameter | typeOfLevel | level | date | time | step | paramId | class | stream | type | experimentVersionNumber |
|---|---|---|---|---|---|---|---|---|---|---|
| q | isobaricInhPa | 100,250,... | 20111215 | 0 | 0,6,... | 133 | od | oper | fc | 0001 |
| t | isobaricInhPa | 100,250,... | 20111215 | 0 | 0,6,... | 130 | od | oper | fc | 0001 |
| tp | surface | 0 | 20111215 | 0 | 0,6,... | 228 | od | oper | fc | 0001 |
| u | isobaricInhPa | 100,250,... | 20111215 | 0 | 0,6,... | 131 | od | oper | fc | 0001 |
| v | isobaricInhPa | 100,250,... | 20111215 | 0 | 0,6,... | 132 | od | oper | fc | 0001 |
| z | isobaricInhPa | 100,250,... | 20111215 | 0 | 0,6,... | 129 | od | oper | fc | 0001 |
OK, let’s select ‘t’ and have a closer look at what it has.
[5]:
fs.describe('t')
[5]:
| shortName | t |
|---|---|
| name | Temperature |
| paramId | 130 |
| units | K |
| typeOfLevel | isobaricInhPa |
| level | 100,250,300,500,700,850,1000 |
| date | 20111215 |
| time | 0 |
| step | 0,6,12,18,24,30,36,42 |
| class | od |
| stream | oper |
| type | fc |
| experimentVersionNumber | 0001 |
We have 8 forecast steps over 7 levels. We will make three selections:
temperature at all fc steps for one level
temperature at all fc steps for three levels
temperature at two fc steps and three levels for a specific date
Filtering with read()
[6]:
# temperature at all fc steps for one level
t_one = mv.read(data=fs, param='t', levelist=500)
t_one.ls()
[6]:
| centre | shortName | typeOfLevel | level | dataDate | dataTime | stepRange | dataType | gridType | |
|---|---|---|---|---|---|---|---|---|---|
| Message | |||||||||
| 0 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 0 | fc | regular_ll |
| 1 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 6 | fc | regular_ll |
| 2 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 12 | fc | regular_ll |
| 3 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 18 | fc | regular_ll |
| 4 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 24 | fc | regular_ll |
| 5 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 30 | fc | regular_ll |
| 6 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 36 | fc | regular_ll |
| 7 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 42 | fc | regular_ll |
[7]:
# temperature at all fc steps for three levels
t_three = mv.read(data=fs, param='t', levelist=[100, 300, 500])
t_three.ls()
[7]:
| centre | shortName | typeOfLevel | level | dataDate | dataTime | stepRange | dataType | gridType | |
|---|---|---|---|---|---|---|---|---|---|
| Message | |||||||||
| 0 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 0 | fc | regular_ll |
| 1 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 0 | fc | regular_ll |
| 2 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 0 | fc | regular_ll |
| 3 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 6 | fc | regular_ll |
| 4 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 6 | fc | regular_ll |
| 5 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 6 | fc | regular_ll |
| 6 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 12 | fc | regular_ll |
| 7 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 12 | fc | regular_ll |
| 8 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 12 | fc | regular_ll |
| 9 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 18 | fc | regular_ll |
| 10 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 18 | fc | regular_ll |
| 11 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 18 | fc | regular_ll |
| 12 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 24 | fc | regular_ll |
| 13 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 24 | fc | regular_ll |
| 14 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 24 | fc | regular_ll |
| 15 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 30 | fc | regular_ll |
| 16 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 30 | fc | regular_ll |
| 17 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 30 | fc | regular_ll |
| 18 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 36 | fc | regular_ll |
| 19 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 36 | fc | regular_ll |
| 20 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 36 | fc | regular_ll |
| 21 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 42 | fc | regular_ll |
| 22 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 42 | fc | regular_ll |
| 23 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 42 | fc | regular_ll |
[8]:
# temperature at two fc steps and three levels for a specific date
t_date = mv.read(data=fs, param='t', levelist=[100, 300, 500], step=[6, 24], date=20111215)
t_date.ls()
[8]:
| centre | shortName | typeOfLevel | level | dataDate | dataTime | stepRange | dataType | gridType | |
|---|---|---|---|---|---|---|---|---|---|
| Message | |||||||||
| 0 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 6 | fc | regular_ll |
| 1 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 6 | fc | regular_ll |
| 2 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 6 | fc | regular_ll |
| 3 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 24 | fc | regular_ll |
| 4 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 24 | fc | regular_ll |
| 5 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 24 | fc | regular_ll |
Filtering with select()
[9]:
# temperature at all fc steps for one level
t_one = fs.select(shortName='t', level=500)
t_one.ls()
[9]:
| centre | shortName | typeOfLevel | level | dataDate | dataTime | stepRange | dataType | gridType | |
|---|---|---|---|---|---|---|---|---|---|
| Message | |||||||||
| 0 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 0 | fc | regular_ll |
| 1 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 6 | fc | regular_ll |
| 2 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 12 | fc | regular_ll |
| 3 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 18 | fc | regular_ll |
| 4 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 24 | fc | regular_ll |
| 5 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 30 | fc | regular_ll |
| 6 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 36 | fc | regular_ll |
| 7 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 42 | fc | regular_ll |
[10]:
# temperature at all fc steps for three levels
# note that this example shows that select() performs some re-ordering of
# the fields for efficiency of filtering - compare with the equivalent read()
# example in the above section
t_three = fs.select(shortName='t', level=[100, 300, 500])
t_three.ls()
[10]:
| centre | shortName | typeOfLevel | level | dataDate | dataTime | stepRange | dataType | gridType | |
|---|---|---|---|---|---|---|---|---|---|
| Message | |||||||||
| 0 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 0 | fc | regular_ll |
| 1 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 0 | fc | regular_ll |
| 2 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 0 | fc | regular_ll |
| 3 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 6 | fc | regular_ll |
| 4 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 6 | fc | regular_ll |
| 5 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 6 | fc | regular_ll |
| 6 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 12 | fc | regular_ll |
| 7 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 12 | fc | regular_ll |
| 8 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 12 | fc | regular_ll |
| 9 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 18 | fc | regular_ll |
| 10 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 18 | fc | regular_ll |
| 11 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 18 | fc | regular_ll |
| 12 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 24 | fc | regular_ll |
| 13 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 24 | fc | regular_ll |
| 14 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 24 | fc | regular_ll |
| 15 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 30 | fc | regular_ll |
| 16 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 30 | fc | regular_ll |
| 17 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 30 | fc | regular_ll |
| 18 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 36 | fc | regular_ll |
| 19 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 36 | fc | regular_ll |
| 20 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 36 | fc | regular_ll |
| 21 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 42 | fc | regular_ll |
| 22 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 42 | fc | regular_ll |
| 23 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 42 | fc | regular_ll |
[11]:
# temperature at two fc steps and three levels for a specific date
t_date = fs.select(shortName='t', level=[100, 300, 500], step=[6, 24], date=20111215)
t_date.ls()
[11]:
| centre | shortName | typeOfLevel | level | dataDate | dataTime | stepRange | dataType | gridType | |
|---|---|---|---|---|---|---|---|---|---|
| Message | |||||||||
| 0 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 6 | fc | regular_ll |
| 1 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 6 | fc | regular_ll |
| 2 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 6 | fc | regular_ll |
| 3 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 24 | fc | regular_ll |
| 4 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 24 | fc | regular_ll |
| 5 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 24 | fc | regular_ll |
[12]:
# alternative: specify date using datetime
import datetime
t_date = fs.select(shortName='t', level=[100, 300, 500], step=[6, 24], date=datetime.datetime(2011, 12, 15, 0, 0))
t_date.ls()
[12]:
| centre | shortName | typeOfLevel | level | dataDate | dataTime | stepRange | dataType | gridType | |
|---|---|---|---|---|---|---|---|---|---|
| Message | |||||||||
| 0 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 6 | fc | regular_ll |
| 1 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 6 | fc | regular_ll |
| 2 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 6 | fc | regular_ll |
| 3 | ecmf | t | isobaricInhPa | 100 | 20111215 | 0 | 24 | fc | regular_ll |
| 4 | ecmf | t | isobaricInhPa | 300 | 20111215 | 0 | 24 | fc | regular_ll |
| 5 | ecmf | t | isobaricInhPa | 500 | 20111215 | 0 | 24 | fc | regular_ll |
Performance
Now we will compare the performance of these two methods and see how they differ when called multiple times on the same data. Imagine that we will want to compute a temporal mean for each level. To do that, we will loop over the levels and select all the forecast steps for each. We will enhance that code by adding timings, and by running the code multiple times, each time taking one more level into account.
[13]:
def filter_n_times(n, use_select):
levs = all_levels[0:n+1]
for lev in levs:
if use_select:
a = fs.select(shortName='t', level=lev).count()
else:
a = mv.read(data=fs, param='t', levelist=lev).count()
# find the list of all levels
t_one_step = fs.select(shortName='t', step=0)
all_levels = t_one_step.grib_get_double('level')
num_levels = len(all_levels)
nlevs = []
times_read = []
times_select = []
for n in range(1, num_levels+1):
nlevs.append(n)
time1 = time.perf_counter()
filter_n_times(n, use_select=True)
timediff = time.perf_counter() - time1
times_select.append(timediff)
time1 = time.perf_counter()
filter_n_times(n, use_select=False)
timediff = time.perf_counter() - time1
times_read.append(timediff)
import matplotlib.pyplot as plt
plt.plot(nlevs, times_select, label="select")
plt.plot(nlevs, times_read, label="read")
plt.xlabel("Number of filters")
plt.ylabel("time (s)")
plt.legend(loc='upper left')
plt.title('select vs read - small values mean faster')
plt.show()
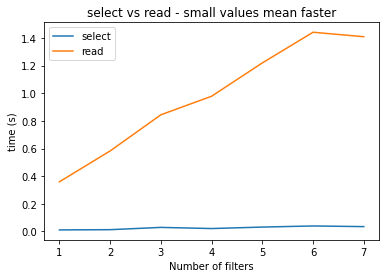
The performance differences are due to the fact that select() caches its Fieldset indexes so that further filtering on the same Fieldset should be faster.